Metagenomics
Through sequencing the 16S rRNA variable region, we can analyze the bacterial flora rapidly and precisely. The sample can be clinical tissue, soil or food.We offer the total solution of service workflow from the DNA extraction to data analysis. You can get a complete report for the metagenomics research. Metagenomics analysis enable researchers to analyzed the intestinal flora, skin flora, and environmental soil flora in the macroscopic scale. Since the NGS technology become more mature due to the longer sequence length and lower error rate, many metagenomics discoveries have been found in several research. Metagenomics has becomes the hottest topic for the researchers.
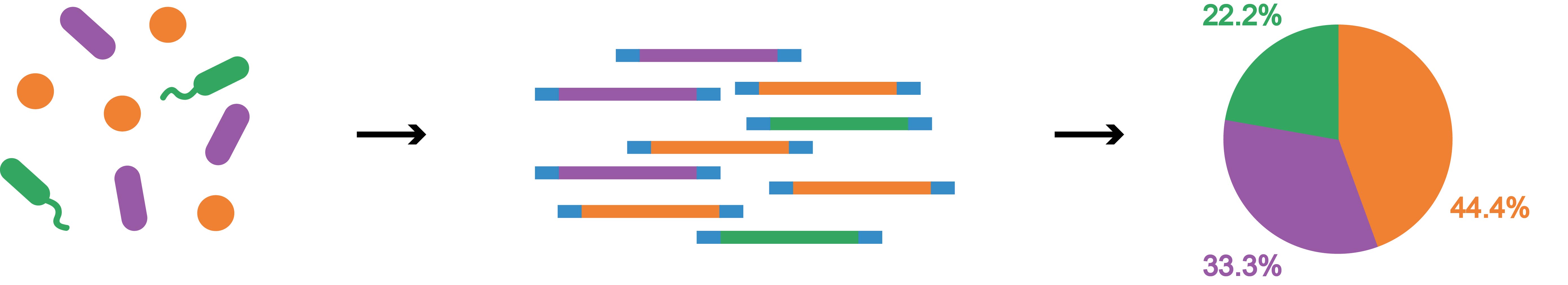
We will offer at least one hundred thousand illumine NGS reads and the bacteria sequence will be classified into different taxonomy scale: kindom, phylum, class, order, family, genus, and species, and so that we can know the composition of the microorganism.
The principles of metagenomics
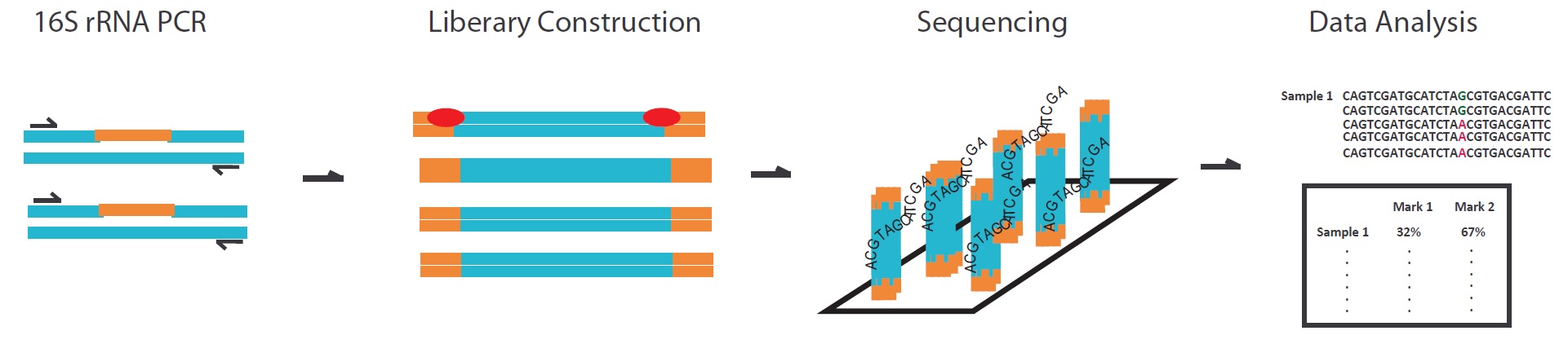
We will amplify the 16s rRNA by the PCR primers which targeted the high conservative region, and then add the index and set up the library. The NGS data will be classified to the bacteria species by RDP Classifier based on the GreenGenes database. The sequence reads can show the content of each bacteria strain.
Service workflow
- Experimental discussion and sample confirmation
- Prepare DNA sample
- Set up DNA library and perform the NGS analysis
- Data analysis and explore the report
The DNA sample quality and input should fit the following requests
- The ratio of OD 260/280 ratio should between 1.8 to 2.
- The concentration of DNA should be 20 ng/ul and the total volume should be more than 20 ul.